Resonant Elastic X-ray Scattering (REXS)
Number of elements in the periodic table (K and L edges)
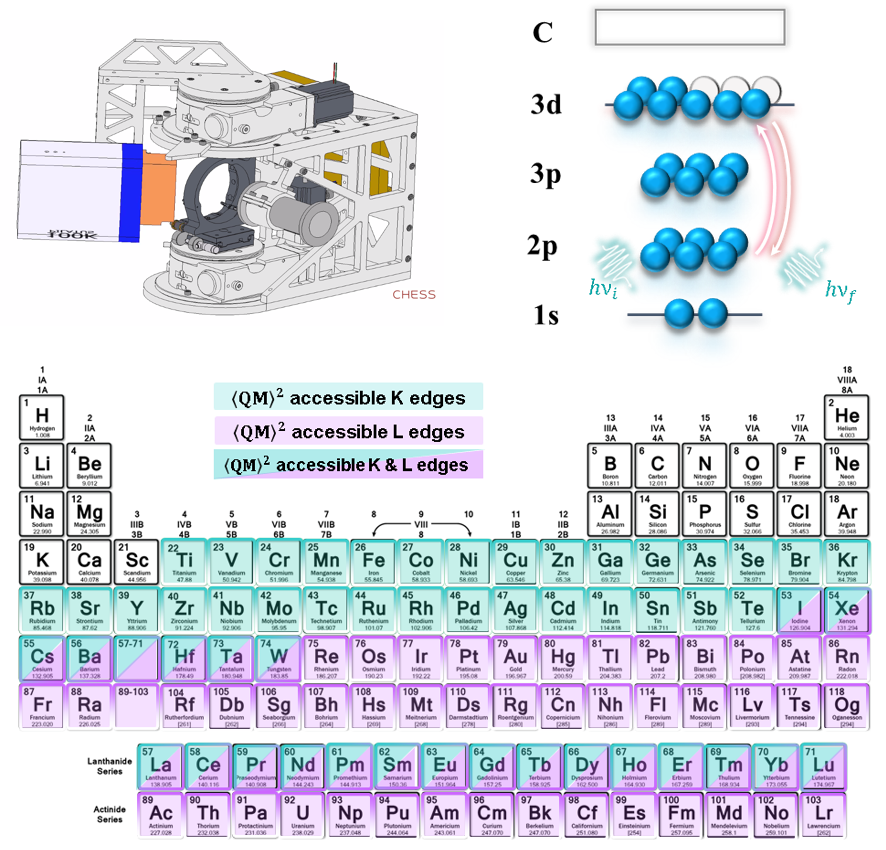
Basic Theory - REXS¶
Basic steps for sample alignments - REXS¶
Beamsplitting polarimeter¶
The polarization analyzer isolates and measures a unique series of diffraction resonances at the different K-edge.
Important SPEC commands for REXS¶
Opening and closing the shutter
opens- Opening the shuttercloses- Closing the shutter
Pilatus video on and off
pil_von- Turn pilatus video 'on'pil_voff- Turn pilatus video 'off'
Position of the displex
wm cryx- where motor for displex in 'x' directionswm cryy- where motor for displex in 'y' directionswm cryy- where motor for displex in 'y' directions
Get the height correct for the sample
opens; lup cryz -0.4 0.4 20 0.2; closes- open the shutter and perform the sample scan in the z directionsumv cryz CEN- it will go to the center position
Get the theta correct for the sample
umv th 10- Move theta to 10 degreesopens; lup cryz -0.4 0.4 20 0.2; closes- open the shutter and perform the sample scan in the z directions
Calulate the motor position in a given point in the reciprocal space
setlat- set lattice paramter information (a, b, c, alpha, beta, gamma)pa- it will show the orientation matrixca- calculate motor settings for a given point in reciprocal spaceca 0 0 20- Calculate the theta, 2theta, chi, and phi for the structural peak (0 0 20)br- br andubr- move to the reciprocal space coordinates (HKL)ubr 0 0 20- Go to the peak (0 0 20)or0 0 0 20- To put the peaks (0 0 20) as first element of the orientation matrixor1 0 0 18- To put the peaks (0 0 18) as first element of the orientation matrix
Moving energy above and below the resonance, perform energyscan
moveE 11.216- move the energy for the sample Ir L edgegetE- Get energymatchUE 3- the undulator 3rd harmonic match with the energylscan L-0.1 L+0.1 50 0.5orlscan 19.9 20.1 50 0.5- Lscan in a range of 0.2, with 50 steps, and 0.5 sec/steptimescan 5- it is a timescan with 1 datapoint in 5 secondsopens; EUQscan 11.2 11.24 40 3 3; closes: Energyscan to get the edge
Moving the energy to the off-resonance
moveE 11.23- Moves to 11.23 keV from current energymatchUE 3- the undulator 3rd harmonic match with the energy, the h k l values will be different than the previous oneubr H K L- the previous h k l values carried by H K L, we need to do ubr H K Lwh- where is every positiongetE- it gies current energy information
We have two detectors: Pilatus 6 (PIL6) and Pilatus 8 (PIL8). PIL6 is in the beam direction. PIL8 is our sigma-pi channel. The alignment will be based on PIL6.
ROI information¶
To set the ROI's for PIL6
getroi6- provide information about the roi chosen in the area detector based on the direction beam position, i.e., xmin, ymin, xsize, ysize.setroi6 278 67 1 3- Changing the vertical and horizontal sizes. Examples: 278 and 67 are minimum pixel dimension along x (vertical) and y (horizontal) axis whereas 1 is vertical pixel size (it is minimum) and 3 is horizontal pixel size.
To set the ROI's for PIL8
getroi- provide information about the roi chosen in the area detector based on the direction beam position, i.e., xmin, ymin, xsize, ysize.setroi 278 67 1 3- Changing the vertical and horizontal sizes. Examples: 278 and 67 are minimum pixel dimension along x (vertical) and y (horizontal) axis whereas 1 is vertical pixel size (it is minimum) and 3 is horizontal pixel size.
After setiing up the two peaks, you can set up the macros to optimize the peak positions
def alignfine '
opens
lup chi -0.2 0.2 50 0.1
umv chi CEN
lup th -0.05 0.05 50 0.1
umv th CEN
lup tth -0.1 0.1 50 0.1
umv tth CEN
lup th -0.03 0.03 30 0.1
umv th CEN
lup tth -0.1 0.1 50 0.1
umv tth CEN
lup chi -0.2 0.2 50 0.1
umv chi CEN
closes
wh
‘
To permform the L scan for (0 1 17) peaks
def getlscan '
wh
opens
lscan 16.96 17.04 120 1
closes
moveE 11.20
matchUE 3
ubr H K L
opens
lscan 16.96 17.04 120 1
closes
moveE 11.215
matchUE 3
‘
Perform two theta plan
def gettthscan2 ‘
wh
alignfine
opens
lup cryz -0.2 0.2 40 0.1
umv cryz CEN
alignfine
opens
th2th -0.04 0.04 80 1
closes
‘